Spatially Expanding
Hierarchical Trees for Compliant Multi-Dimensional Scaling
Thomas Rebotier,
Interactive Cognition Laboratory,
University of California, San Diego
Abstract:
This article suggests a new type of initial configuration to use
for gradient descent multidimensional scaling algorithms such as the Kruskal-Shepard
algorithm. Using a binary hierarchical
tree of the points to scale, one can expand that tree in the final space,
starting with the root and repeatedly replacing each node by its two
successors; at each expansion one uses the gradient descent again to reshape
the configuration. Compared to applying
gradient descent to the result of classical scaling, tree expansion yields similar
levels of stress, but renders better the grouping of points belonging to
similar clusters.
Introduction
When trying to visualize the proximity structure of a high
dimensional pattern, one frequently has to choose between clustering into a
hierarchical tree, or scaling down to two or three dimensions. Conventional wisdom considers that
multi-dimensional scaling (MDS) is good at representing the big picture,
whereas hierarchical clustering (HC) handles
local details more accurately (Kruskal 1977; Arabie, Hubert et
al. 1996).
Ideally, one would want a low-dimensional configuration rendering both
the global and local properties; such that, for example, if one drew between the
final points a hierarchical tree obtained from the original data, this tree
would appear simple and related branches would stay next to each other. This paper suggests a method that finds such
tree-friendly scaling configurations.
There is a second situation where existing strategies come short:
if there exists extraneous information in addition to the dissimilarities. For example, the points can be extracted from
known different populations, such as male and female participants in an
experiment, or different political parties in an opinion poll. The different clusters may or not have the
same spatial distribution; yet of the many stress-equivalent ways of
representing the data, some must better than others at representing this
extraneous information. Likely, it is
better to group together points belonging to the same cluster. In some cases, the preexisting classification
may be hierarchical. For example, a
library can display a 2-dimensional map of topics and sciences by scaling
similarities between sciences; in which case it is important to respect the
library’s call number system along with the similarity structure. Until now, both classical and non-metric
scaling are ill-equipped to represent jointly the similarity structure and
extraneous information. In contrast, the
method suggested here can use such extraneous information.
Before we introduce our method, let us review the shortcomings of
current MDS strategies. Consider a set
of data points (Pi) of known dissimilarities (δi,j)
that has to be visualized in two dimensions.
When the dissimilarities should approximate directly actual distances
between points, the MDS is metric,
and can be done in two ways: by classical MDS, in which the points are projected
orthogonally in the subspace encoding a maximum of variance (Young and Householder 1938;
Torgerson 1952; Cox and Cox 2001), or by a metric gradient descent,
trying to minimize the sum of residuals (the squared differences between
distances and the corresponding dissimilarities). When the mapping between dissimilarities and
distances is unknown—for example obscured by an unknown psychometric function—one
can rely only on the order of the
dissimilarities, and scaling is achieved by repeating a gradient descent trying
to minimize the sum of residuals between distances and the same distances
monotonically reduced to the dissimilarities (Shepard 1962; Shepard 1962;
Kruskal 1964; Kruskal 1964).
In the following paragraphs we will examine in turn why classical,
metric MDS misrepresents smaller inter-point distances, and how gradient
descent algorithms, including non-metric scaling, depend on initial
configurations.
Why
existing scaling strategies deal poorly with local structure:
1. In the case of classical multi-dimensional scaling (CMDS),
the (δi,j)’s are taken to represent
Euclidian distances, and (Pi) can be rendered after a
Young-Torgerson reduction, by projecting the points in two dimensions for
graphing (Young and Householder 1938;
Torgerson 1952; Cox and Cox 2001).
However, CMDS is not the best least squares rendering of distances– it
is only the best rendering by orthogonal projection. In fact the sum of residuals (sum of squared
error over inter-point distances) computed for a CMDS result is always high, in
part because CMDS, like all orthogonal projections, systematically decreases
distances. The following example (Figure
1) shows where problems can lie:
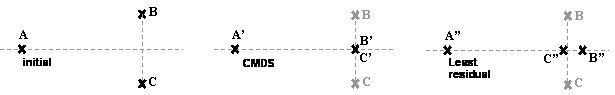
Figure 1:
Classic MDS will project B and C onto the same point at intersection of
dotted axes, but minimal least square
error for the distances within a one-dimensional configuration is for B” and C”,
whose center is at dAB from A, and whose distance dB”C” =
dBC/3.
Figure 1
illustrates two problems:
- Because it lies parallel to an
eigenvector pruned by the CMDS, the distance between B and C has been ignored. In contrast, the distance between A and
the other points is almost conserved, because it is almost collinear to
the eigenvector preserved by the CMDS.
- The projection ignores the arc,
so that even though B and C are both at the same distance from A, their
projection gets closer than that distance.
A first step in estimating the true efficiency of CMDS is to
dilate resulting distances by a factor ρ2 = Σij [(δij.dij)/(dij2)]. This factor makes up for dividing the average
arc by its cosine. Even then the sum of
residuals can easily lag by 20% behind that obtained after a gradient descent
adjustment (see the results section and Figure 5).
Still, CMDS reduces the original distances unequally, depending on
which eigenvector bears most of a distance.
For this reason, CMDS can mistreat gravely the details of local
structure when these details lie in dimensions that are suppressed. When the algorithm deals with
high-dimensional underlying patterns, it is very likely that many local
patterns will be entirely misrepresented because of this CMDS shortcoming. So, if one wants to render as accurately as
possible the set of between-point distances, CMDS projection must be followed
by a gradient descent to minimize the sum of square errors.
However, even metric gradient descent starting from the CMDS
configuration can fail to properly restitute local sub-patterns, because when
the initial projection brings inside a 2-D sub-pattern points that should lie
outside, the gradient descent is unable to move them outside, and the
“pressure” caused by their presence distorts local details. This is illustrated below in Figure 2, where
CMDS superimposes two clusters that should be kept separated, and gradient
descent as a result explodes one of the clusters into a ring around the other
one.
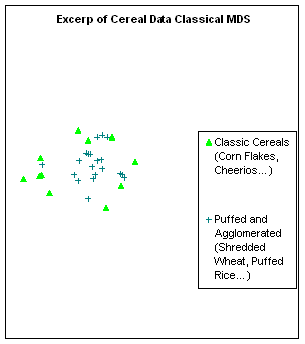
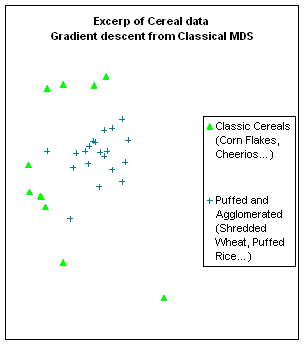
Figure 2: Detail of MDS made on the ASA Cereal
data set. Two out of seven types of
cereals are projected on top of each other by CMDS. Applying gradient descent worsens this
particular feature of the configuration, because the central repulsion creates
a crown of local minima in which the “classic cereal” points settle.
2. In the non-metric case, the (δij) are on an ordinal rather than a
ratio scale, in which case an initial configuration is modified to optimize the
rank order of the resulting distances, (di,j). The Shepard-Kruskal algorithm (Shepard 1962; Shepard 1962;
Kruskal 1964; Kruskal 1964) establishes for existing distances
(dij) a set of target distances (d’ij) that are in the
same order as the (δij)’s—when
dij is already in order, d’ij is set to equal dij, and
when sequences of dij ’s are in disarray, all corresponding d’ij ’s are set
to the average of these dij,’s.
The target distances are then used as dissimilarities in a metric
gradient descent that modifies the initial configuration to minimize Stress
(the normalized sum of residuals, Σ(dij-d’ij)2/Σdij2 ). Because this metric gradient descent can
disturb the rank order again, the process must be iterated, until the
configuration is stable.
Because gradient descent adjusts an initial configuration to a
local minimum, its result depends on this initial
configuration. To obtain a good final
solution, one possible strategy is to try several random initial configurations
and keep the most favorable. While this
method in theory can explore the entire space of solutions, it is
computationally costly. In practice a
few attempts will generally yield at least one solution with acceptable stress.
A second strategy is to use the result of CMDS as a starting configuration;
this can be done after applying CMDS either on the dissimilarities themselves
(treating them as distances), or on pseudo-distances reconstructed to be
consistent with the rank order of dissimilarities (Lingoes and Roskam 1973).
This latter strategy is a one-shot attempt, computationally effective
but final. It can occasionally fail, as
illustrated in Figure 3, and cannot be improved gracefully by increasing
computational power. In conclusion, neither
random configurations nor a CMDS-produced configuration are ideal starting
points for the Kruskal-Shepard non-metric MDS.
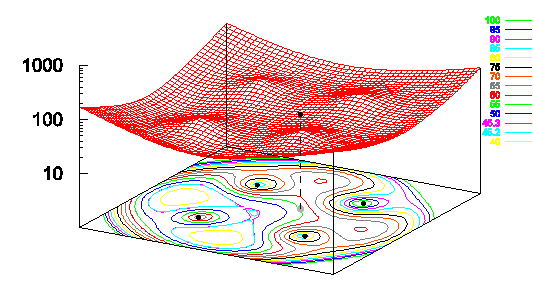
Figure 3: Local minima in a metric energy
landscape and failure of CMDS-seeded gradient descent. When scaling the five points (4, 4, 0),
(4,-4,0), (-4,4,0), (-4,-4,0) and (1,1,7), CMDS projects the latter in the
plane Z=0, onto the point (1,1,0) shown in grey. This graph shows a 2-D section of a
10-variable function: the sum of residuals landscape, by fixing the coordinates
of the 4 points that were already in the plane (“the base”) and showing the sum
of residuals for different locations of the 5th point. For moving the grey point only, there are at
least five local minima: four located outside the base and one about (-1, -1,
0). In fact, this latter point is where
gradient descent takes the grey point, but it is not the absolute minimum.
The
tree-expansion strategy:
In summary, gradient descent appears as the best method to render
the dissimilarities, but on one hand it requires a good starting configuration,
and on the other hand it does not take in consideration the supplementary information
that can be given by a hierarchical tree.
We suggest solving these two problems together by using that
supplementary information to construct a starting configuration which gradient
descent will optimize with regard to stress.
In the process of arranging the points (Pi) in two
dimensions, the dissimilarity structure is unavoidably impoverished – but it
can be impoverished less, by keeping in the final configuration some of the
information obtained by a clustering algorithm, independently of the scaling
algorithm.
In order to jointly minimize stress and render the cluster
structure, we propose to apply gradient descent (or Kruskal-Shepard) repeatedly
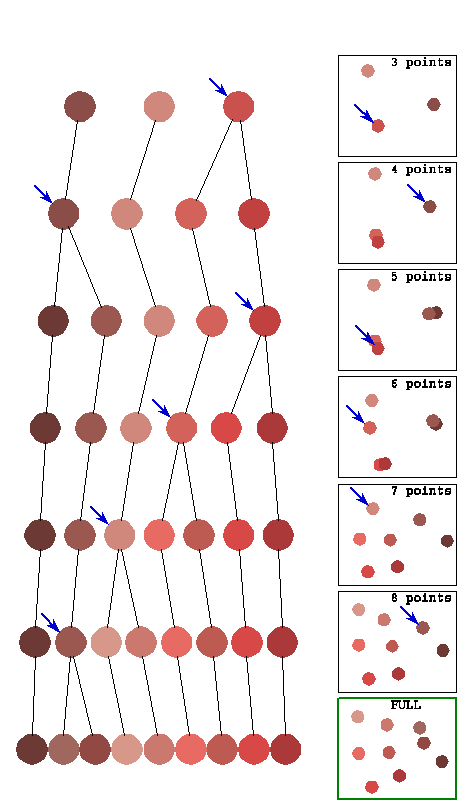
to a growing configuration, obtained by expanding the hierarchical tree. This is illustrated by Figure 4 in which the
hierarchical tree and successive configurations are shown bide by side. Prior to using this algorithm, one needs the
dissimilarity matrix at all stages of expansion. If the tree has been constructed from a SAHN
algorithm these dissimilarities are already available, otherwise they have to
be computed in a one-sweep forward pass.
Given these dissimilarities, the algorithm is to:
1.
Position
a starting configuration:
a.
Take a section of the hierarchical tree
containing D+1 points where D is the final dimension (e.g., 3 points for a
planar MDS). This is the current section of the tree.
b. Using
the dissimilarities between these points, place them in space exactly using
CMDS.
c.
Test that the resulting configuration
defines a variety of dimension D. If
yes, this is the starting configuration.
If no, use as current section a
section with one more point (the highest numbered tree node in the current
section is replaced by its two offspring), and go to step b.
2.
Expand the tree:
a.
In the current
section, select the highest tree node, and replace it, in place, by its
offspring;
b. Adjust
the resulting pattern by gradient descent (or Kruskal-Shepard) to a “decent”
accuracy.
i.
One wants to adjust the location of all
existing points only to a precision Δ, just more accurate than the
movements we can expect to happen at the next stage of the algorithm. The idea is to avoid finessing the
convergence past an accuracy that will be undone anyway by the next split. Because Δ must be computed before the
convergence, on estimates of the next positions, it is necessarily
approximate. After testing several heuristics,
we have settled for taking Δ = (1/16).(δP’P”2/max(δij)2. The gradient descent is stopped by a test on
the relative movement: when for all points their movement divided by the
average distance between points is smaller than Δ.
ii.
In applying gradient descent, one has the
choice to take into account the mass of the points or not:
1.
Points are considered of equal mass, SSE= Σij
(dij-δij)2
2.
Heavier points move less, SSE= Σij
mimj (dij-δij)2
The
results presented use the weighted algorithm, number 2.
c.
Return to step a. until all point in the
current configuration represent terminal nodes.
3.
Make
a final adjustment down to the desired accuracy.
The algorithm allows the initial configuration to grow from the hierarchical
organization of the rendered pattern. Points
spawned by the same node stay near that starting point, and insofar as dissimilarities
allow they will remain in the same region throughout the expanding process. Thus, points that should be clustered
together are not initially separated by a gradient “mountain pass”, as they
often would be in a random initial configuration. This algorithm shares with others the
benefits of gradient descent, but with respect to the local optimum issue it improves
over other methods by finding a local optimum that respects as much as possible
the hierarchical tree.
Simulations
The goal of simulations was to validate the algorithm and start
estimating its strengths and weaknesses.
Too many variables were involved to chart the complete problem space;
the two most important that were not considered were variations in the type of
classification tree, and in configuration dimension. The variables that were
varied involve the number of points and different conditions of grouping. We present side-by-side results from the
metric gradient descent and from non-metric, Kruskal-Shepard gradient
descent.
Simulation
Method:
N patterns of P points were randomly generated in RP-1: either all from the same
population, with each coordinate from a random Gaussian distribution of mean
zero and variance one, or from C different populations, deviating with variance
one from cluster centroids themselves drawn randomly with a different variance
(the parameter “clusterscale”). Random
deviates were computed using the routine “gasdev()” from (Press, Teukolsky et al. 1992). After generation, each pattern was centered on
zero. For the non-metric simulations,
the distances dij these were transformed into dissimilarities δij by the following function:
δij=
dij + .49* sin(2*dij) + noise*gasdev() ; in which noise=0.1 unless reported otherwise.
Each pattern was then organized as a binary ordered hierarchical
tree by the centroid-based SAHN algorithm (Gordon 1996), and the tree was used for our
expansion algorithm. In the non-metric
case, we built the SAHN tree from the dissimilarities. Because the dissimilarities were noised
distances, they no longer conformed to the triangular inequality. As a result, when using constructing the SAHN
tree directly from dissimilarities, negative square dissimilarities would
occasionally appear. One way to solve
this would be to use some of the additive constant methods developed prior to
non-metric scaling. However we found it
more efficient to simply adjust the computed dissimilarity during the SAHN
algorithm by the following function:
δ’ = δ+ + e-δ2 in which δ+ is the positive part of δ.
When testing for the ability to render known clusters, the tree
was computed with a forced compliance to the original clusters so that each of
the different original populations corresponds exactly to a sub-tree. This was done during the agglomeration phase
of SAHN by choosing the two closest points under the condition that they
belonged to the same cluster, up to step (N-C) of agglomeration, at which stage
nodes were in one-to-one correspondence to the original clusters, and the
agglomeration could be finished without constraint. This type of tree is hereafter called
“cluster-compliant”, or simply “compliant” tree.
In the metric simulations the gradient descent was made on the sum
of residuals, SSE = Σij
(dij-δij)2. In the non-metric simulations, following
Kruskal, the gradient descent was made on the stress computed with distances
monotonically regressed to dissimilarities:
Stress = ( Σ mimj(dij-d’ij)2 )
/ ( Σ mimj(dij)2
)
in which (d’ij) are the
distances regressed monotonically to the similarities and mi is the
mass of node i.
For the metric program, we did not adopt such a complicated step
length computation as Kruskal’s; instead we monitored 3 consecutive steps and
decreased the step length if these were increasing or oscillating, and
increased it if they were decreasing too fast.
In conjunction with this simpler procedure, and for computing time
reasons, we automatically cancelled trials for which the convergence was too
long for any one of the gradient descents.
The upper limit was arbitrarily set at 100*N3; when a trial
was cancelled its results were discarded for all methods together. Quite possibly this pruning of difficult
configurations has introduced a small bias, however for N>10 very few trials
were cancelled, and because we are reporting medians rather than means we do
not believe that the results are much affected.
For the non-metric simulations we followed Kruskal’s formula for the
step length.
This data was then used to find fitting configurations by the
following methods:
- Classic
Multi-Dimensional Scaling (CMDS).
- CMDS
rescaled by a dilatation or ratio Σij [(δij.dij)/(dij2)]
(for minimal residuals).
- CMDS
plus dilatation followed by a gradient descent.
- Expanding
the SAHN-tree without considering mass.
- Expanding
the SAHN-tree, considering mass.
- Expanding
the compliant tree without considering mass.
- Expanding
the compliant tree, considering mass.
- Doing
gradient descent from circular initial conditions (all initial points equally
spaced on a circle).
- Doing
gradient descent from random initial conditions.
Considering the mass during expansions had a very minor effect, so
the Figures report only results from weighted expansion. Similarly, circular
initial conditions results are almost identical with random initial condition
results. We will most often report only
the latter. Each number reported is the median of 99 trials using independent
random patterns. We also recorded, but
found no interest in reporting, the mean, and the lowest and highest 5%
quantiles.
Simulation
results:
Comparing Stress from different
methods: Figure 5.
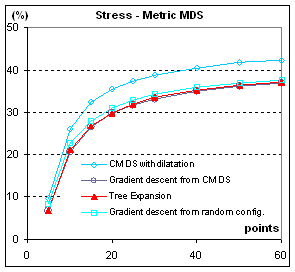
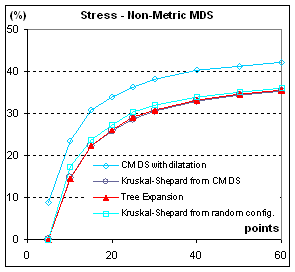
Figure 5: Stress, in %, of configurations from
different MDS methods. S= Σij (δij-dij)2 / Σij dij2. All points are from the
same population (C=1). When parameters
are varied (C>1, clusterscale from 0.0 to 4.0), the stress diminishes when
clusters are better separated and augments when there are more clusters with
the same separation, but the different methods stay in the same ratio.
Rendering Contingent Data Structure:
Figures 6 and 7.
To test how good each MDS algorithm is at rendering accidental
structure appearing in data and captured by a SAHN tree, we measured how well
clusters defined by the SAHN tree are separated in the final
configuration. Using patterns of N2
points we adopt the N clusters defined by sectioning the tree at the
appropriate level. (Because the tree
groups points on the basis of their observed distance, the clusters can contain
more or less than N points; they are only constrained to an average of N point
per cluster.) The chosen measure of how
well these clusters are separated in the configuration obtained by one of the
scaling algorithms is the Fisher significance of the groups they define. I.e., from the coordinates of the points in
the final configuration, an F-ratio can be computed, and from it a
p-value. This p-value reflects the
probability that a random configuration would keep the clustered points as well
together as the observed configuration does.
The p-value is not a perfect measure, in particular it improves when the
points that belong to the same cluster get closer to each other, regardless of
what their dissimilarity is. By over-gathering
the points, one gets a better p-value at the expense of a higher stress; notably,
Classical Multi-Dimensional Scaling does just that, because it tends to squash
more strongly smaller dissimilarities, which happen to lie more often within
the clusters than between them. However,
for solutions equivalent in stress, the p-value reflects how well the clusters
are separated.
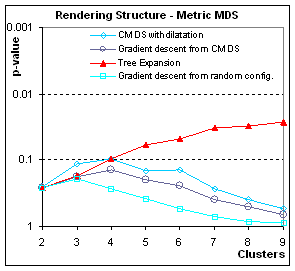
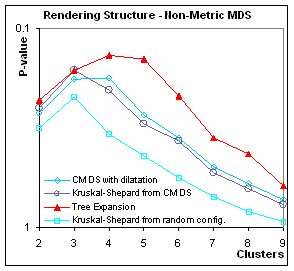
Figure 6:
How well is contingent structure rendered? These graphs plot p-value (the significance
of the grouping) in function of the number N of clusters (for a total of N2
points). When using metric scaling
(gradient descent on the dissimilarities), the accidental organization of
homogeneous data (all points come from the same Gaussian distribution) is can
be preserved by Tree Expansion, but not when other algorithms are used. When using non-metric scaling, Tree Expansion
still fares better than other methods, but overall all methods fail to preserve
structure.
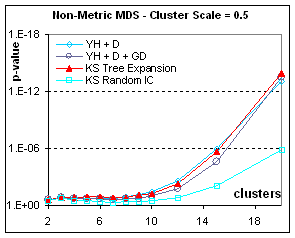
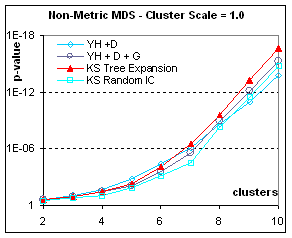
Figure 7: Rendering contingent structure of
heterogeneous data: if the data is drawn from different populations, accidental
structure is more pronounced and the p-value becomes more significant. Here we show only the non-metric MDS results
(metric results are similar). The more
separated the population the easier it is to get good final separation from any
kind of initial condition; however tree expansion is always the best initial
condition. When the scale is only 0.5,
good separation only appears for numerous configurations (more than 100
points). With very heterogeneous data
(clusters separated by more than one bandwidth), Classical MDS (in this case, applied to
dissimilarities) can occasionally give the best p-value, not because it
separates clusters better, but because in the same distance-squashing process
that causes a higher stress, CMDS brings points of the same cluster closer to
their centroid. In all cases, Tree
expansion gives a better p-value than Kruskal-Shepard gradient descent from any
other initial condition.
Rendering pre-assigned clusters:
Figure 8.
Sometimes the grouping of data points is known in advance from
extraneous data. For example, clustering
could represent the value of a between-subject factor, observed or
assigned. In this case the tree-expansion
algorithm must be used with a compliant
tree, that is, a tree that first organizes the data points inside clusters
before it organizes the clusters. With
such a tree, expansion will first arrange spatially cluster centroids, then
arrange the individual points making up these clusters.
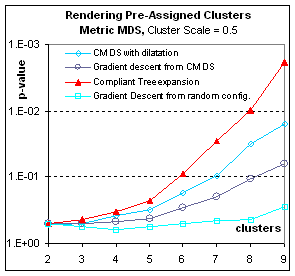
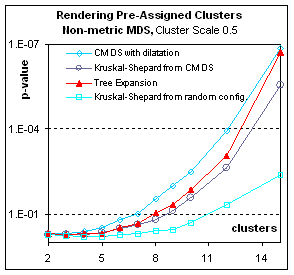
Figure 8: How well are pre-assigned clusters
rendered? These graphs plot p-value (the significance of the
grouping) in function of the number N of clusters (for a total of N2
points). Here the pre-assigned clusters
correspond to different populations in the full-dimensional space, such that
the centroids of the clusters are distributed with 0.5 times the bandwidth used
to distribute points inside clusters around their centroid. The clusters are therefore widely
overlapping. Nevertheless, both in the
metric and the non-metric case, Multidimensional scaling can render in two
dimensions the difference between populations.
Expanding the compliant tree is of all gradient descents the most
efficient way to render the clusters (In fact it eventually surpasses CMDS in
spite of the p-value bias for the latter, at 20 clusters – not shown; p-values
for more than 20 clusters could not be computed as they exceeded the double
float precision but the F statistic indicated that eventually gradient descent
from CMDS also outperforms simple CMDS).
If the clusters do not correspond to any difference in spatial
repartition (Result not shown) – Cluster Scale=0.0, corresponding to a
homogenous population with arbitrary labeling– all p-values are very poor (over
.5) but compliant tree expansion is the best of those bad solutions.
Computational Cost: Figure 9.
One of the worries one might have with the tree expansion method
is about the cost of repeating the work of repeatedly organizing almost the
same configuration. The results
presented in Figure 9 show that this worry is partly justified. For non-metric scaling, tree expansion is
about 2.5 times as slow as doing Kruskal-Shepard only once, from the results of
CMDS. This does not take into account
the lower order costs of building the tree for one algorithm, or of
diagonalizing the double-centered dissimilarity matrix for the other algorithm. For metric scaling, tree expansion is
actually the cheapest way. This result
is counter-intuitive if one thinks in terms of re-doing the work, but makes
sense when considering that each expansion stage provides a starting
configuration very close to the final configuration for that stage.
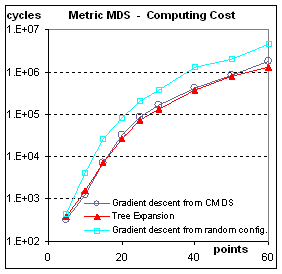
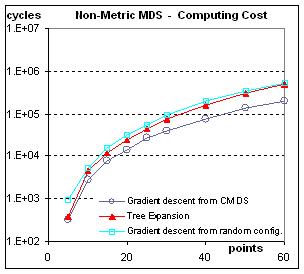
Figure 9: Computational Cost. The cost is measured in basic cycles (computing
the contribution of one pair of points to one coordinate of the gradient). Even though it is related to the
implementation, this cycle count allows to compare the speed of convergence
from different initial conditions.
Cost of doing better than Tree
Expansion:
If the main focus is on the quality of the configuration obtained,
it makes sense to consider the strategy of starting gradient descent repeatedly
from many random initial conditions. One
may hope that through the force of sheer numbers, a solution will be found that
simultaneously lowers stress and renders well the original accidental
structure. Tables 1 and 2 present very
contrasting results, denying this hope for metric scaling, but keeping the door
open for non-metric scaling. Each column
presents data from 99 configurations for each of which results from 100 random
initial conditions were compared to the result of tree expansion. We show the mean percentage of random initial
conditions doing better than tree expansion on stress only, p-value only, and
on both simultaneously. To give an idea
of the distribution we also show the median configuration and the 5th
centile most favorable to random initial conditions.
|
Metric MDS
%age of RIC trials as good or
better than Tree Expansion…
|
Points in Configuration
|
5
|
10
|
15
|
20
|
25
|
30
|
40
|
|
Clusters
|
2
|
3
|
4
|
5
|
6
|
6
|
7
|
|
…for Stress only
|
mean %
|
69*
|
26*
|
21
|
16
|
13
|
12
|
8
|
|
…for p-value only
|
mean %
|
75*
|
42*
|
17
|
9
|
3
|
3
|
1
|
|
…for both Stress and p-value simultaneously
|
mean %
median
%
5thcentile %
|
15
0
69
|
3
0
30
|
0
0
0
|
0
0
0
|
0
0
0
|
0
0
0
|
0
0
0
|
|
Non-Metric MDS
%age of RIC trials as good or better than Tree Expansion…
|
Points in Configuration
|
5
|
10
|
15
|
20
|
25
|
30
|
40
|
50
|
60
|
80
|
|
Clusters
|
2
|
3
|
4
|
5
|
6
|
6
|
7
|
7
|
8
|
9
|
|
… for Stress only
|
mean %
|
76
|
32
|
32
|
31
|
37
|
31
|
31
|
28
|
25
|
26
|
|
… for p-value only
|
mean %
|
34
|
40
|
38
|
27
|
21
|
25
|
17
|
22
|
18
|
18
|
|
…for both Stress and p-value simultaneously
|
mean %
median
%
5thcentile %
|
15
4
59
|
4
0
37
|
6
0
41
|
4
0
53
|
3
0
19
|
3
0
35
|
3
0
28
|
4
0
37
|
2
0
30
|
4
0
48
|
Tables 1 and 2: Comparing Random Initial Condition
trials to Tree Expansion. The mean and
highest 20-ile of 99 data points, where each time 100 trials were run with
different initial configurations and their result compared to the result of
tree expansion. In the “both
simultaneously” case, we report the mean rather than the median, and also
report the 5th centile, because floor effects quickly intervene: the
median for “both” is zero for all number of points. The 5th centile is the number of
random trials, out of 100, doing better than tree expansion in the one-in-twenty
situation where that number is the highest.
Another remarkable fact is that the probabilities for a random trial of
being better than tree expansion for stress, and for p-value, are not
independent, but inversely related. This
shows that tree expansion strikes a remarkable compromise between both
criteria.
* For small
configurations the large majority of random initial configuration converges to
the same configuration as tree expansion does.
Comments on tables 1 and 2:
- For
non-metric MDS, random initial conditions results are more frequently
equal to or better than tree expansion results. We believe that it is because Non-metric
MDS is less demanding than metric MDS (it only tries to respect the order
of the original dissimilarities) and that therefore the process of
organizing intermediary stages of expansion is stunted by being stopped as
soon as intermediary dissimilarities are “good enough”.
- For
metric MDS, percentages of random initial conditions that yield as good a
result on either stress or p-value steadily decrease with the number of
points in a pattern.
- For
non-metric MDS, the percentage of random initial conditions that yield as
good a result on either stress or p-value is almost constant, and confirm
common wisdom that 4 or 5 random trials are usually enough to get a good
value of stress.
- For
both metric and non-metric MDS, the distribution of results is very
skewed. The medians are much lower
than the means; i.e., there are a few of the 99 configurations for which
many random conditions will do fairly well, but for the large majority
they do very poorly.
- For
metric MDS, the probabilities of being better on Stress and being better
on p-value are not independent; if they had been the mean on both would be
a product of the means on each. In
fact the percentage of simultaneous success is much lower than can be
predicted from the percentages of being better on one count only. This suggests that for metric MDS there
has to be a trade-off between both properties. For this trade-off, both tree expansion
and gradient descent from classical MDS strike a much better compromise
than random conditions can hope to reach; within that compromise, tree
expansion favors p-value and descent from classical MDS favors stress.
- For
non-metric MDS, the probabilities of being better on Stress and being
better on p-value seem to be fairly independent.
Example:
Multidimensional Scaling of Cereal data:
As an example, several scaling
techniques were applied to the data offered for the 1993 ASA Statistical
Graphics Exposition (http://lib.stat.cmu.edu/datasets/1993.expo/cereal). The data has 13 fields, the first two being
categorical: cereal manufacturer, and hot or cold cereal. “Hot or cold” was encoded as +1 or -1. Thereafter, all 12 numerical fields were
normalized to a mean 0 and a variance 1, and between-cereal distance was
computed in the resulting 12-dimensional Euclidian space. Two approaches were taken to this data:
first, by clustering according to the cereal manufacturer, and second, by
building a SAHN Tree (centroid-based).
Clustering by manufacturer did not yield any significant classification
– p values for all methods were greater than .5. For the second approach, all scaling methods
were able to cluster the points decently, but tree expansion did so much
better. Table 3 summarizes the
quantitative results, and Figure 10 shows the resulting configurations.
|
|
Metric
|
Non-Metric (Kruskal-Shepard)
|
|
Stress
|
p-value
|
cost index
|
Stress
|
p-value
|
Cost index
|
|
Tree Expansion
|
23.8 %
|
3.9 e-6
|
127,769
|
18.4 %
|
1.7 e-6
|
561,814
|
|
CMDS + D with GD or KS
|
23.4 %
|
1.8 e-3
|
335,566
|
18.5 %
|
1.1 e-5
|
154,154
|
|
CMDS + D without gradient
descent
or Kruskal-Shepard descent.
|
32.7 %
|
2.0 e-4
|
n/a
|
29.8 %
|
2.4 e-4
|
n/a
|
|
non weighted Tree Expansion
|
23.2 %
|
1.9 e-4
|
498,118
|
18.4 %
|
1.9 e-6
|
692,216
|
|
Circular Initial Configuration
(better than random on most
counts)
|
24.0 %
|
1.9 e-3
|
1,555,708
|
20.2 %
|
2.2 e-3
|
492,107
|
Table 3: Stress, p-value, and computing cost for scaling configurations
of the cereal data, obtained ether by Tree Expansion or by gradient descent (or
Kruskal-Shepard) completing CMDS.
|
Symbol
|
Ad-hoc title
and description
|
Cereals in that
cluster
|
|
|

|
Fruits and Nuts
fairly rich in protein and
fat
but low in slow carbs
|
Almond_Delight R
Crispy_Wheat_&_Raisins G
Life Q
Quaker_Oat_Squares Q
Clusters G
Raisin_Nut_Bran G
|
Cracklin'_Oat_Bran K
Great_Grains_Pecan P
Muesli_Raisins,_Dates,_&_Almonds
R
Muesli_Raisins,_Peaches,_&_Pecans
R
100%_Natural_Bran Q
|
|

|
Fruits and Bran
heavier – densest cereals
|
Just_Right_Fruit_&_Nut K
Total_Raisin_Bran G
Post_Nat._Raisin_Bran P
Raisin_Bran K
Fruitful_Bran K
|
Fruit_&_Fibre_Dates,_Walnuts,_and_Oats
P
Oatmeal_Raisin_Crisp G
Basic_4 G
Nutri-Grain_Almond-Raisin K
Mueslix_Crispy_Blend K
|
|
|
Classic Cereals
high slow-carb content,
hi sodium
|
Just_Right_Crunchy__Nuggets
K
Total_Corn_Flakes G
Total_Whole_Grain G
Product_19 K
Cheerios G
|
Special_K K
Corn_Chex R
Rice_Krispies K
Corn_Flakes K
Rice_Chex R
Kix G
|
|
x
|
Puffed and agglomerated
low fat, sugar and fiber
content,
medium slow carb content
|
Puffed_Rice Q
Puffed_Wheat Q
Frosted_Mini-Wheats K
Strawberry_Fruit_Wheats N
Raisin_Squares K
Shredded_Wheat_'n'Bran N
Shredded_Wheat_spoon_size N
Shredded_Wheat N
Multi-Grain_Cheerios G
Wheaties G
|
Bran_Chex R
Wheat_Chex R
Crispix K
Triples G
Double_Chex R
Nutri-grain_Wheat K
Grape_Nuts_Flakes P
Bran_Flakes P
Grape-Nuts P
|
|
*
|
Hot cereals
|
Cream_of_Wheat_(Quick) N
Maypo A
Quaker_Oatmeal Q
|
|
|

|
Pure Bran
low calories, low slow
carbs,
low sugar, high potassium
|
100%_Bran N
All-Bran K
All-Bran_with_Extra_Fiber K
|
|
|
+
|
Fancy cereals
more sugar,
lower shelf
placement
|
_Crisp P
Smacks K
Apple_Jacks K
Corn_Pops K
Froot_Loops K
Lucky_Charms G
Cocoa_Puffs G
Count_Chocula G
Trix G
Fruity_Pebbles P
|
Honey-comb P
Cap'n'Crunch Q
Honey_Graham_Ohs Q
Cinnamon_Toast_Crunch G
Apple_Cinnamon_Cheerios G
Honey_Nut_Cheerios G
Nut&Honey_Crunch K
Wheaties_Honey_Gold G
Golden_Grahams G
Frosted_Flakes K
|
Table 4: The
clusters defined by a section of the SAHN tree at the 7th
level. The cluster titles are descriptive,
and the comments below the titles summarize the main common properties,
observed manually, for the cluster members.
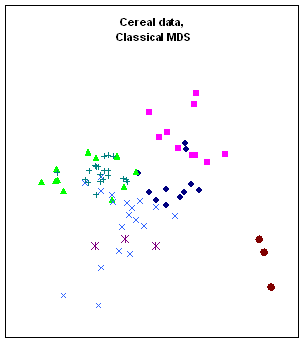
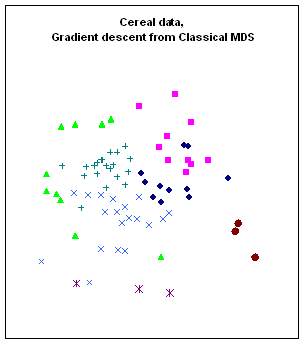
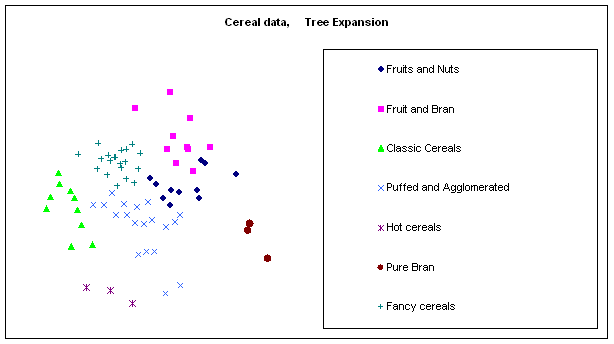
Figure 10:
Rendering the cereal data by different algorithms: a. classical scaling (Young-Torgerson
reduction); b. classical scaling followed by a gradient descent to minimize
residual square error; c. expansion of the SAHN tree, applying gradient descent
to adjust the configuration every time a point is split. Notice how Classic cereals (rendered by green
triangles) are dispersed when gradient descent takes classical scaling as an
initial configuration, because that cluster overlaps another one which is more
compact. This is a “mountain pass” effect, as is probably the isolation of two
“dark diamond” points inside the cluster of “purple squares”. In contrast, the tree expansion is relatively
free of such effects.
Ramifications:
The computations described here have just begun to explore a large
set of possible variants. Only one type of tree was used (binary SAHN centroid
tree). It is likely that different types
of trees are more efficient for different data sets. Tree expansion does not have to be restricted
to expanding binary trees; one can easily figure extensions of the method using
any kind of tree. When computing time is
not critical, we suggest expanding several candidate trees.
In fact expanding a non-binary tree is equivalent to skipping a
few stages of intermediary gradient descent in expanding a binary tree. Such a strategy can be employed deliberately
if computing power is at stake, or simply if it is known from the structure of
the data that fewer stages of expansion are sufficient. The archetypical example would in the
situation of rendering known clusters, to first use tree expansion (or another
algorithm) to place the cluster centroids, and then in one stage replace all
centroids by the points of the corresponding cluster.
One variant we considered and which occasionally yielded better
results than reported for the main variant was not to consider the differences in
centroid mass during the gradient descents.
The simulations were limited to 2-D configurations, even though
the algorithm applies also for uni-dimensional scaling (UDS). The uniqueness of UDS stems from two
peculiarities: first, the local minima problems of gradient descent are much
worse in one dimension, and second, being in one dimension only facilitates a
combinatorial approach (Defays 1978).
We believe that for UDS, the best use of tree expansion will require its
combination with heuristic combinatorial techniques. The simplest example is to confine heuristic
exploration to the group of permutations along the tree (the group generated by
all transpositions of two direct offspring of the same node), which is a
subgroup of only 2N out of the N! permutations of all points. For large data sets, one could explore all
tree-defined permutations only a few levels down from the current level. For example, when expanding from four point
to five, rather than select the immediate best configuration of 5 points (best
out of two choices), one could consider all 16 potentially resulting
configurations of 8 points, then finalize the choice of the five point
configuration to the configuration leading to the lowest 8–point stress. In the application of tree expansion, as in
other matters, UDS deserves a fully independent treatment, because of the facility
with which it allows combinatorial schemes, and because of the connections
between UDS and tree-building.
Even for 2-D or 3-D configurations, tree expansion can be applied
in conjunction with some random exploration of initial conditions space. For example, one can replace the early part
of expansion, placing the first P points by random exploration, and then expanding
those nodes into a full configuration by normal tree expansion. The configuration obtained by tree expansion
at various steps can be used as one of the strains for a genetic
algorithm—in other problems requiring
gradient descent, genetic algorithms (Goldberg 1989) have been efficient ways of
cutting down the complexity of random exploration (Nolfi, Elman et al. 1994).
Tree expansion can also be combined with heuristic search by
applying the heuristic at the level of tree-building. For a tree built bottom-up (as SAHN trees
are), some agglomeration choices are obvious and some are not. By combining all the close calls into a family
of alternate trees, one creates as many possible ways of seeking an optimal
configuration.
Finally, Tree Expansion may be combined with other strategies that
can bias the rendering in favor of particular clusters. The most prominent of these is to weight the
contribution of different pairs of points in the gradient, in order to favor
either the within-cluster or the between-cluster distances. Because the good properties of tree expansion
originate from setting a proper initial configuration, all others methods that
do not rely on particular initial conditions should benefit from being used in
conjunction with tree expansion.
Summary
and conclusion:
This article suggests a new type of initial configuration to use
for gradient descent multidimensional scaling algorithms such as
Kruskal-Shepard. Using a binary
hierarchical tree of the points to scale, one can expand that tree in the final
space, starting with the root and repeatedly replacing each node by its two
successors; at each expansion one uses the gradient descent again to reshape
the configuration. Compared to applying
gradient descent to the result of classical scaling, tree expansion is tidier,
and more apt at keeping together points belonging to the same cluster. In particular, when the clusters tend to
overlap, tree expansion can find significant representations whereas other
methods fail.
Besides our main result, the explorations made in this paper shed
light on basic differences between metric and non-metric MDS, that users should
keep in mind when they have a choice between both methods. Metric MDS is more demanding; by doing
gradient descent on the dissimilarities themselves it strives for
perfection. In comparison, non-metric
MDS only strives for distances in the right order. In the scope of our simulations, these
differences twice caused finer results from metric MDS: first in that
contingent structure of homogeneous data can be rendered significantly by
metric MDS (Figure 6), but not by non-metric; and secondly, in the trade-off
that metric MDS forces between finding a good stress and rendering structure
(Table 1). The goal of the
Kruskal-Shepard algorithm is as much finding the psychometric function relating
dissimilarities to distances as it is to find those distances. To make the Kruskal-Shepard algorithm as
demanding on the distances as metric MDS is, one would need to add constraints
on the shape of the psychometric function, constraints that would make further
demands than the simple monotonous regression.
In fact, there lies a continuum of algorithms from metric MDS, where the
psychometric function is imposed to be identity, to the Kruskal-Shepard
algorithm, where it can take any monotonous shape. For this reason, the choice between metric
and non-metric MDS –or other intermediary algorithms—should be dictated not by
convenience, but by what one expects the psychometric function to be.
The results presented here should be taken as the floor, not the
ceiling, of tree expansion efficiency.
Firstly, there is no guarantee that type of tree used here was the best for
the testing data. Secondly, the data might
not be the most difficult for the classical methods that we compared tree
expansion to. In our results, metric
scaling of the dissimilarities followed by gradient descent fare quite well,
always better than random initial conditions.
This is not the case for all possible problems; for example Arabie and
Boorman (1973, p.160) report scaling configuration
representing partitions for which CMS followed by non-metric gradient descent
fared worse than random initial conditions.
In contrast to the metric scaling of similarities, tree expansion is a
robust process, which we expect to degrade well with problem difficulty, its
weakness being only the representative quality of the tree.
By using tree expansion, classical metric and non-metric scaling
becomes a field of application of hierarchical clustering, and the large
existent corpus of clustering algorithms must be tested to find out which tree
are the best candidates for expansion.
Acknowledgements:
The author wishes to thank for their help Professor David Kirsh,
Daniel Bauer, and John Hershey. This
work was funded by a grant from France Telecom.
Reference:
Arabie, P. and S. A. Boorman (1973).
"Multidimensional scaling of measures of distance between
partitions." Journal of Mathematical Psychology 10: 148-203.
Arabie, P., L. J. Hubert, et al.
(1996). Clustering and Classification. Singapore, World Scientific.
Cox, T. F. and M. A. A. Cox (2001).
Multidimensional Scaling. Boca Raton, Chapman & Hall.
Defays, D. (1978). "A short
note on a method of seriation." British Journal of Mathematical
Statistical Psychology 31:
49-53.
Goldberg, D. E. (1989). Genetic
Algorithms, in search, Optimization and Machine Learning. Readin, Mass.,
Addison-Wesley.
Gordon, A. P. (1996). Hierarchical
Classification. Clustering and Classification. P. Arabie, L. J. Hubert
and G. de Soete. Singapore, World Scientific. 1: 65-121.
Kruskal, J. B. (1964).
"Multidimensional Scaling by Optimizing Goodness of Fit to a Nonmetric
Hypothesis." Psychometrika 29(1):
1-27.
Kruskal, J. B. (1964).
"Nonmetric Multidimensional Scaling: a Numerical Method." Psychometrika
29(2): 115-129.
Kruskal, J. B. (1977). The
relationship between Multidimensional Scaling and Clustering. Classification
and clustering. J. van Rezin. Reading, MA, Addison-Wesley: 17-44.
Lingoes, J. C. and E. E. Roskam
(1973). "A mathematical and empirical study of two multidimensional
scaling algorithms." Psychometrika Monograph Supplement 38.
Nolfi, S., J. L. Elman, et al.
(1994). "Learning and evolution in neural networks." Adaptive
Behavior 3(1): 5-28.
Press, W., S. Teukolsky, et al.
(1992). "Numerical Recipes in C, second edition."
Shepard, R. (1958). "Stimulus
and response generalisation: tests of a model relating generalization to
distance in psychological space." Journal of Experimental Psychology
55(6): 509-523.
Shepard, R. N. (1962). "The
Analysis of Proximities: Multidimensional Scaling with an Unknown Distance
Function. I." Psychometrika 27(2):
125-140.
Shepard, R. N. (1962). "The
Analysis of Proximities: Multidimensional Scaling with an Unknown Distance
Function. II." Psychometrika 27(3):
219-246.
Torgerson, W. S. (1952).
"Multidimensional scaling: 1. Theory and method." Psychometrika
17: 401-419.
Young, G. and A. S. Householder
(1938). "Discussion of a Set of Points in Terms of Their Mutual
Distances." Psychometrika 3(1):
19-22.